-

-
-
-
-
-
-
-
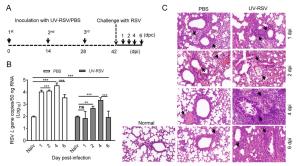
dpi Groups Histopathology scorea Alveolar tissue Peribronchial spaces Perivascular spaces 1 PBS 2.38 ± 0.24 1.93 ± 0.36 1.92 ± 0.33 UV-RSV 2.62 ± 0.12 2.02 ± 0.12 1.97 ± 0.23 2 PBS 1.98 ± 0.33 1.72 ± 0.27 1.72 ± 0.37 UV-RSV 2.50 ± 0.28 2.02 ± 0.17 1.98 ± 0.20 4 PBS 1.42 ± 0.13 1.48 ± 0.21 1.48 ± 0.16 UV-RSV 3.55 ± 0.25 2.78 ± 0.19 2.73 ± 0.20 6 PBS 1.60 ± 0.17 1.62 ± 0.17 1.50 ± 0.18 UV-RSV 3.17 ± 0.47 2.50 ± 0.34 2.45 ± 0.34 aThe histological scores were blindly evaluated by the degree of inflammation in alveolar tissue, peribronchial and perivascular spaces. Scores ranged from 0 (normal) to 3 or 4 (severe), as described in Materials and Methods. Data represent the mean ± SD (n = 6). Table 1. Histopathology score of lungs in PBS-treated and UVRSV vaccinated mice after RSV challenge.
-
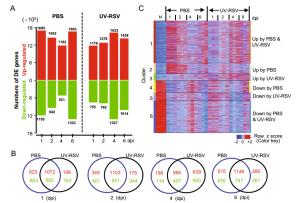
Clusters Gene count Representative GO functional enrichmenta (P value) Cluster 1 2078 Immune system process, inflammatory response, immune response, response to virus, cell cycle, apoptotic process, antigen processing and presentation, TNF signaling pathway Cluster 2 521 Inflammatory response, immune system process, PI3K-Akt signaling pathway, TNF signaling pathway Cluster 3 344 Extracellular exosome, membrane, peptidase activity Cluster 4 594 Cilium movement, cell projection, Cluster 5 389 Angiogenesis, cell surface, calcium ion binding Cluster 6 1584 Cell adhesion, cilium movement, angiogenesis, multicellular organism development, cell surface, cell junction, Rap1 signaling pathway, Calcium signaling pathway, cAMP signaling pathway aRepresentative Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) categories are enriched by hypergeometric (FDR-adjusted P value < 0.05). Table S4 in the supplemental material for all enriched GO and KEGG categories and their corresponding P values. Table 2. Representative GO and KEGG functional enrichment of six clusters.
-
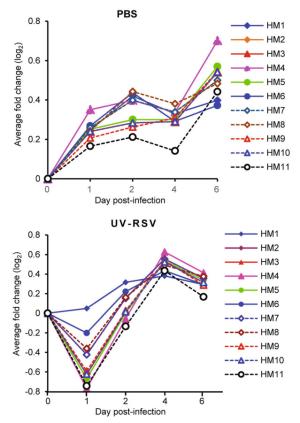
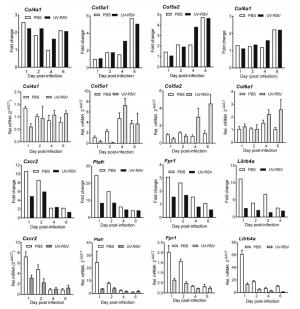
HMs Gene counts Representative GO functional enrichmenta (P value) Representative genes HM 1 237 Cell cycle, DNA replication, cellular response to DNA damage stimulus, microtubule based movement Cdk1, MCM7, MCM8, BRCA1, CHAF1B, TICRR HM 2 53 Protein ubiquitination, intracellular signal transduction, ubiquitin protein ligase activity, ubiquitin mediated proteolysis UBE2L6, KLHL13, SOCS3, SOCS1, SIAH1B PTGER4, GLP1R, PTGIR, CALCRL, PTGER2, GCGR, ADRB1 HM 3 41 Adenylate cyclase-activating G-protein coupled receptor signaling pathway, positive regulation of camp biosynthetic process, G-protein coupled receptor signaling pathway, camp-mediated signaling, neuroactive ligand-receptor interaction, vascular smooth muscle contraction, calcium signaling pathway Ly6k, Cpm, Lypd8, Cntn4, Vnn1, Vnn3 HM 4 28 Anchored component of membrane, cell adhesion molecules (cams), pantothenate and coa biosynthesis Col5a1, Col5a2, Col1a1 Col1a2, Col4a1, Col4a2, Col6a1 HM 5 39 Collagen fibril organization, cell adhesion, cellular response to amino acid stimulus, extracellular matrix structural constituent, metal ion binding, protein digestion and absorption, ECM-receptor interaction, PI3K-Akt signaling pathway Ifih1, Ifit3, Ifit1, Dhx58, Mx1, Dhx58 HM 6 78 Defense response to virus, negative regulation of viral genome replication, immune system process, innate immune response, double-stranded RNA binding, Influenza A, herpes simplex infection Avpr1a, Adra1b, Bdkrb1, Chrm3, Mchr1, P2ry1, P2ry6 HM 7 53 Signal transduction, G-protein coupled receptor signaling pathway, positive regulation of cytosolic calcium ion concentration, neuroactive ligand-receptor interaction, calcium signaling pathway H2-T23, H2-M3, Fcgr4, CD14, Tap2, Tap1, H2-T3 HM 8 61 Antigen processing and presentation of peptide antigen via MHC class I, immune system process, adaptive immune response, peptide antigen binding, receptor binding, phagosome, graft-versus-host disease Slc11a1, Cd59a, Cd59b, Cyba, Cd53, Cybb HM 9 44 Positive regulation of phagocytosis, cell surface receptor signaling pathway, respiratory burst, inflammatory response, complement and coagulation cascades Gsdmd, B2m, Orm2, Orm3, Pglyrp1 HM 10 41 Defense response to Gram-positive bacterium, acute-phase response, immune system process Psma5, Psma4, Psmb2, Psmb8, Psmb9, Psmb10 HM 11 37 Antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent, proteolysis involved in cellular protein catabolic process, proteolysis, threonine-type endopeptidase activity, peptidase activity, proteasome, NF-kappa B signaling pathway Psma7 aRepresentative GO and Kyoto Encyclopedia of Genes and Genomes (KEGG)categories are enriched by hypergeometric (FDR-adjusted P value < 0.05). Table S6 in the supplemental material for all enriched GO and KEGG categories and their corresponding P values. Table 3. The representative GO and KEGG functional enrichment of eleven conserved high-influential modules.
Figure 7 个
Table 3 个