HTML
-
COVID-19 is caused by SARS-CoV-2, a novel beta coronavirus and now affecting more than 210 countries, areas or territories (WHO 2020; Zhou et al. 2020; Zhu et al. 2020). The World Health Organization (WHO) has declared the COVID-19 outbreak as a global pandemic. SARS-CoV-2 is highly contagious with an estimated R0 of 2–3 and causes COVID-19 with a wide range of clinical manifestations including asymptomatic infection to severe pneumonia with deadly outcome (Hellewell et al. 2020; Tian et al. 2020; Wang et al. 2020; Zhao et al. 2020). According to different reports published so far, COVID-19 has a mortality rate of 2%–10% (Li et al. 2020b; Yang et al. 2020a).
COVID-19 has an incubation period of 2–14 days (Jiang et al. 2020). In the early phase of the clinical course, the clinical manifestations include fever, dry cough, feeling weak, diarrhea, etc., which are rather unspecific and indistinguishable from the common colds. The asymptomatic carriers of SARS-CoV-2 may shed viruses and could be very contagious (Nishiura et al. 2020). Therefore, a highly sensitive and specific laboratory method is essential for early diagnosis of SARS-CoV-2 infection, which is critical for appropriate management of suspected COVID-19 patients.
With abundant sequence information of SARS-CoV-2 strains, many commercial kits for detection of SARS-CoV-2 RNA have been developed (Chan et al. 2020; Chen et al. 2020b; Won et al. 2020). However, unsatisfactory sensitivity of nucleic acid detection kits in clinical application has been reported repeatedly (Li et al. 2020a; Xie et al. 2020). Conflicting positive and negative test results could be obtained from the same patients at different time points along the clinical course (Yang et al. 2020b). In fact, a sequential change of positive-to-negative-to-positive results could not be properly explained but could cause great concern about potential viral reactivation and re-infection in cured COVID-19 patients (Chen et al. 2020a; Ye et al. 2020).
The molecular diagnosis of COVID-19 by RT-PCR detection of SARS-CoV-2 RNAs has been flawed by the problem of false negative results. As any other PCR-based diagnostic method, many factors such as the quality of test kits and specimens may reduce the sensitivity and specificity of the test. An in-depth investigation is needed to identify the major causes for false negative results. In this study, we tried to gain insights by retrospectively analyzing test results and data from carefully matched patient specimens and sequential specimens from patients with conflicting test results.
Our analyses demonstrated that clinical specimens with high cellular contents, measured by the RNA levels of a housekeeping gene ribonuclease P/MRP subunit p30 (RPP30), provided a significantly higher positive rate in SARS-CoV-2 detection. In fact, the detectability of SARS-CoV-2 RNAs in respiratory tract specimens was found to be highly associated with cellular content, represented by RPP30 RNA levels. Based on data from these specimens, a set of Ct cutoff values for RPP30 RT-PCR reaction was established for specific tests based on amplification of specific target regions using specific types of specimens. These Ct cutoff values from RPP30 RT-PCR could be used as criteria for identification of false negative results. Our data also suggest that false negative results accounted for most contradicting test results in SARS-CoV-2 RNA detection in clinical use due to poor specimen quality. Thus, the quality of specimens, defined by the cellular content, is a key factor for satisfactory RT-PCR results and could be assessed by an internal reference such as RPP30.
-
A total of 161 confirmed COVID-19 patients admitted into the Second Hospital of Fuyang from January 20 to February 29, 2020 were enrolled in this retrospective single-center study. The diagnosis of COVID-19 for patients was performed according to the Guidelines for the Diagnosis and Treatment of New Coronavirus Pneumonia (version 7) published by the National Health Commission of China. The study was approved by the Ethics Committee of the Second Hospital of Fuyang (Number 2020020004). Written informed consents were obtained from all patients.
A total of 573 sputum and 579 throat swab specimens were collected from COVID-19 patients. Among them, 254 pairs of matched sputum/throat swab specimens were collected on the same days from the same patients. 109 sputum specimens were collected at different time points from 15 patients with alternating positive–negative–positive results. 72 throat swab specimens from 10 patients were collected in the same way.
-
Laboratory confirmation of the SARS-CoV-2 infection was performed by real-time RT-PCR (Bojie Ltd, Shanghai, China) according to the approval of Chinese Center for Disease Control and Prevention. The fluorescent quantitative RT-PCR tests use two primer pairs and corresponding probes within the ORF1ab and N region of the SARS-CoV-2 genome, respectively (Zou et al. 2020). The oligo DNA sequences for the ORF 1ab were: Forward primer (F)-CCTGTGGGTTTTACACTTAA; Reverse primer (R)-ACGATTGTGCATCAGCTGA; Probe (P)-5′-FAM-CCGTCTGCGGTATGTGGAAAGGTTATGG-BHQ1-3′. For the N region: Forward primer (F)-GGGGAA CTTCTCCTGCTAGAAT; Reverse primer (R)-CAGACATTTTGCTCTCAAGCTG; Probe (P):5′-FAM-TTGCTGCTGCTTGACAGATT-TAMRA-3′. A cellular housekeeping gene ribonuclease P/MRP subunit p30 (RPP30) was used as a marker of the presence of cellular materials and as an internal reference. The sequences for the primer pair and probe were: Forward primer (F)-5′-AGATTTGGACCTGCGAGCG-3′ ' (nt 28–46), Reverse primer (R)-5′-GAGCGGCTGTCTCCACAAGT-3′ (nt 92–73), and the probe (P)-5′-TTCTGACCTGAAGGCTCTGCGCG-3′ (nt 49–71), designed according to the reference sequence NM_006413.5 in the GenBank. A cutoff for the positivity for RPP30 RT-PCR is set at a Ct value = 38.
Sputum and/or throat-swab specimens were collected from patients and maintained in viral-transport medium prior to laboratory testing. The RNA was extracted with a Nucleic Acid Extraction Kit (Shuoshi Ltd, Taizhou, China), The RT-PCR reactions for all three gene fragments were conducted in the same reaction tube on a SLAN 96P Fluorescence Quantitative PCR Amplification Instrument (Hongshi Ltd, Shanghai, China) by following the manual of the kit (Bojie Ltd, Shanghai, China). As recommended by the Guidelines for Diagnosis and Treatment of the New Coronavirus Pneumonia published by the National Health Commission of China, a test result was considered positive if the Ct values of both target region amplifications of SARS-CoV-2 genome in one test were lower than 38, or the Ct value in one of two target region amplifications was lower than 38 in two consecutive tests.
-
Categorical variables are expressed in frequency or percentage. Nonparametric variables were expressed in median and quartile intervals and the significance was tested by Mann–Whitney U or Kruskal–Wallis test. The correlation between two indicators was analyzed by the Pearson methods. The diagnostic values of selected parameters for differentiating negative and positive of SARS-CoV-2 RNA with samples were assessed by receiver operating characteristic (ROC) and the area under the ROC curve (AUC). A P value < 0.05 is considered statistically significant in all statistical analyses. The SPSS statistical software (Macintosh version 26.0, IBM, Armonk, NY, USA) and R package were used for statistical analysis.
Patients and Specimens
RT-PCR Assays
Statistical Analyses
-
SARS-CoV-2 RT-PCR may fail to provide a positive result with specimens from COVID-19 patients. Sputum and throat swabs specimens are commonly used for RNA extraction and RT-PCR detection of SARS-CoV-2 RNA. A retrospective analysis showed that only 39.1% of 573 sputum specimens and 19.2% of 579 throat specimens from COVID-19 patients collected at different time points were consistently tested positive for SARS-CoV-2 RNAs by RT-PCR (Fig. 1A, 1B). Many samples were positive for only the reaction detecting the N region of SARS-CoV-2 RNA. Taking advantage of access to data for a large number of specimens from a cohort of 161 confirmed patients, we systematically analyzed the data to gain insights into the causes for questionable results and find the right solutions.

Figure 1. The RT-PCR detection of SARS-CoV-2 RNAs in 1152 samples from patients with confirmed SARS-CoV-2 infection. 573 Sputum (A) or 579 throat swab (B) specimens were collected and subjected to SARS-CoV-2 specific RT-PCR assays targeting the ORF1ab and N regions. +: RT-PCR positive; −: RT-PCR negative. RT-PCR was performed to detect RPP30 RNAs that reflect the presence of cellular RNAs in sputum (C) and throat swab (D) specimens. +/+: positive results for both ORF1ab and N-specific RT-PCR assays; ± : only one of both RT-PCR assays was positive; −/−: both RT-PCR assays were negative. The negative RT-PCR results of SARS-CoV-2 detection correspond to high Ct values for the RT-PCR for RPP30 RNAs. The results were tested for significance by using the Kruskal–Wallis test (C, D). A P value of < 0.05 is considered as significant.
It is not yet known if viral RNA in released viral particles or in infected epithelial cells or both are detected in these specimens. Thus, we retrospectively examined the presence of cellular materials in patient specimens by detecting the RNA of a cellular housekeeping gene RPP30. RPP30 RNA was frequently detected in sputum and throat swab specimens but at very different levels (Fig. 1C, 1D). In fact, the Ct value of RPP30 RNA ranged from to 18 to 27 for 573 sputum specimens and from 20 to 30 for 579 throat swab specimens (Fig. 1C, 1D). Strikingly, 224 sputum and 111 throat swab specimens positive for SARS-CoV-2 RT-PCR (positive for both ORF1ab and N) have significantly lower Ct values for RPP30 RT-PCR (corresponding to higher levels of RPP30 RNAs) than those with only single positive or double negative results. This fact indicates a strong association between the RPP30 and SARS-CoV-2 RNA levels in patient samples, irrespective of the types of specimens or the target regions of the viral genome for amplification.
-
In order to determine the relationship among amplification of target regions ORF1ab, N region and cellular gene RPP30, data for a total of 254 paired sputum and throat swab specimens were selected and subjected to statistical analyses. In this cohort of specimens, 35.8% of sputum specimens and 16.1% of throat swab specimens were tested positive for SARS-CoV-2 RNA (Fig. 2A, 2B). The correlation between the Ct values of ORF1ab/N-region and RPP30 RT-PCRs was analyzed by using the Pearson method. Consistently, a strong correlation between the Ct values for ORF1ab and N (r = 0.913, P < 0.0001 for sputum, r = 0.926, P < 0.0001 for throat swab), ORF1ab and RPP30 (r = 0.809, P < 0.0001 for sputum, r = 0.805, P < 0.0001 for throat swab), N and RPP30 (r = 0.939, P < 0.0001 for sputum, r = 0.920, P < 0.0001 for throat swab) was found (Fig. 2C–2H). Thus, RPP30 may serve as an internal reference and could be a suitable indicator for the quality of specimens. The same conclusion could be reached when the analysis was performed by excluding specimens for which negative results for SARS-CoV-2 RNA detection were obtained (Supplementary Figure S1).
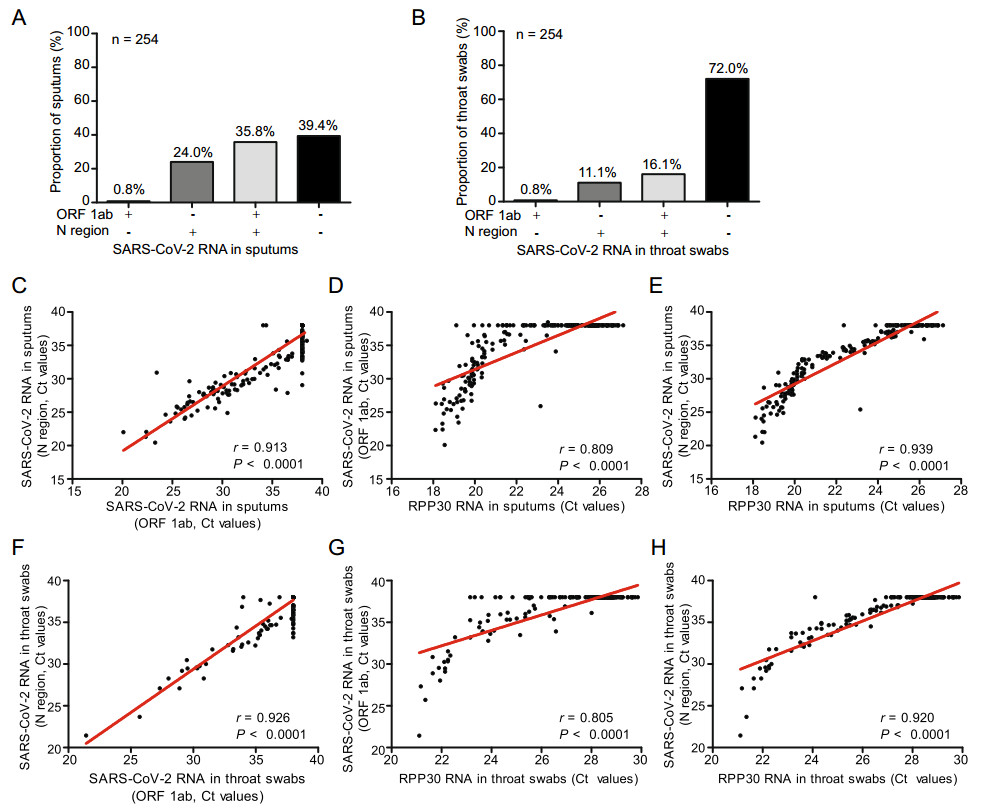
Figure 2. The RT-PCR detection of SARS-CoV-2 RNAs in 254 paired sputum and throat swab specimens from patients with confirmed SARS-CoV-2 infection. 254 paired sputum (A) and throat swab (B) specimens were collected and subjected to SARS-CoV-2 specific RT-PCR assays targeting the ORF1ab and N regions. +: RT-PCR positive; −: RT-PCR negative. Pearson correlation coefficients for the levels of detected SARS-CoV-2 RNA levels (based on the assays for ORF1ab and N region) and RPP30 RNAs in patient samples were calculated. For sputum specimens: C ORF1ab and N region; D ORF1ab and RPP30; E N region and RPP30. For throat swab specimens: F ORF1ab and N region; G ORF1ab and RPP30; H N region and RPP30. A P value of < 0.05 is considered as significant.
-
Having discovered strong associations between amplification of target regions and RPP30, we hoped to identify appropriate Ct values for RPP30 which could be used as cutoffs for distinguishing valid test results from invalid ones. Using the receiver operating characteristic (ROC) and area under the ROC curve (AUC) analyses, the values of RPP30 RNA levels for the identification of false negative results of SARS-CoV-2 RNA detection were assessed. As is shown in Fig. 3A–3D, the sensitivity and specificity could reach 95.03%–95.26% and 83.72%–98.55% respectively for ORF1ab RT-PCR with sputum (AUC 0.981, cutoff 21.53) and throat swab (AUC 0.973, cutoff 25.53) specimens, as well as for the N region-specific RT-PCR with sputum (AUC 0.981, cutoff 24.47) and throat swab (AUC 0.993, cutoff 27.48) specimens.

Figure 3. The suitability of the Ct values of RPP30 specific RT-PCR to discriminate false negative results of RT-PCR detection of SARS-CoV-2 RNAs. The RT-PCR results for SARS-CoV-2 and RPP30 RNAs obtained from 254 paired sputum and throat swab specimens from patients with confirmed SARS-CoV-2 infection were used for analysis. The ROC curve and AUC were calculated for the Ct values of RPP30 specific RT-PCR to predict the false negative results of the ORF1ab and N-specific RT-PCR assays for SARS-CoV-2 RNA detection by using the R package "pROC". The cut offs of the Ct values of RPP30 RT-PCR were set at the sensitivity around 95% for each case: 21.53 for (A) ORF1ab specific RT-PCR with sputum; 24.47 for (B) N specific RT-PCR with sputum; 25.53 for (C) ORF1ab specific RT-PCR with throat swab; and 27.48 for (D) N specific RT-PCR with throat swab. Cr, Criterion (Cutoff value); Se, Sensitivity; Sp, Specificity; A P value of < 0.05 is considered as significant. The Ct values of the RPP30 RT-PCR were used to analyze the positive and negative results of ORF1ab and N-specific RT-PCR assays with 254 paired sputum (E) and throat swab (F) specimens. Pos: positive results; Neg: negative results. The negative RT-PCR results of SARS-CoV-2 detection correspond to high Ct values for RT-PCR for RPP30 RNAs. Red lines: cutoffs for ORF1ab RT-PCR; blue lines: cutoffs for N specific RT-PCR. The results were tested for significance by using the Mann–Whitney test (E, F). A P value of < 0.05 is considered as significant.
To validate the performance of these cutoff values, these cutoff values were applied to data for 253 specimens from patients with conflicting positive and negative test results and for that from patients with multiple samples and data for 254 pairs of sputum and throat swab specimens from confirmed COVID-19 patients, respectively. The prediction results clearly showed that the majority of false results was correspondent to Ct values over the set cut off values for RPP30 RT-PCR irrespective of types of specimens or target regions of RT-PCR reactions (Fig. 3E, 3F).
We then tried to determine whether inadequate cellular content was the cause of conflicting RT-PCR results. Analyses of 181 serial samples from 20 patients with conflicting RT-PCR results showed, similar to larger cohort of samples analyzed above, that 47.7% of sputum specimens and 20.8% of throat swab specimens were tested positive in SARS-CoV-2 RNA testing (Fig. 4A, 4B). Again, Pearson analyses revealed strong correlations between the Ct values of RT-PCR assays for ORF1ab and N (r = 0.921, P < 0.0001 for sputum, r = 0.883, P < 0.0001 for throat swab), ORF1ab and RPP30 (r = 0.870, P < 0.0001 for sputum, r = 0.802, P < 0.0001 for throat swab), N and RPP30 (r = 0.930, P < 0.0001 for sputum, r = 0.929, P < 0.0001 for throat swab) (Fig. 4C–4H).

Figure 4. The RT-PCR detection of SARS-CoV-2 RNAs in 181 sputum and throat swab specimens from 20 patients with conflicting RT-PCR results. 109 paired sputum (A) and 72 throat swab (B) specimens were collected and subjected to SARS-CoV-2 specific RT-PCR assays targeting the ORF1ab and N regions. +: RT-PCR positive; −: RT-PCR negative. Pearson correlation coefficients for the levels of detected SARS-CoV-2 RNA (based on the assays for ORF1ab and N region) and RPP30 RNAs in patient samples were calculated. For sputum specimens: C ORF1ab and N region; D ORF1ab and RPP30; E N region and RPP30. For throat swab specimens: F ORF1ab and N region; G ORF1ab and RPP30; H N region and RPP30. The Ct values of RPP30 RT-PCR were used to analyze the positive and negative results of ORF1ab and N-specific RT-PCR assays with 109 sputum (I) and 72 throat swab (J) specimens. Pos: positive results; Neg: negative results. The negative RT-PCR results of SARS-CoV-2 detection correspond to high Ct values for RT-PCR for RPP30 RNAs. Red lines: cutoffs for ORF1ab RT-PCR; blue lines: cutoffs for N specific RT-PCR. The results were tested for significance by using the Mann–Whitney test (I, J). A P value of < 0.05 is considered as significant.
We further examined the relationship between cellular content and validity of test results by applying the RPP30 cutoff values established above to data from a cohort of specimens for which contradicting results had been obtained. As is shown in Fig. 4I, 52 positive and 57 negative results were obtained from the ORF1ab RT-PCR reactions with sputum specimens while 84 positive and 25 negative results were generated from the N-specific RT-PCR reactions. Of specimens with negative results, the great majority had RPP30 Ct values above the cutoffs established (Fig. 4I). Similarly, 15 positive and 57 negative results for ORF1ab RT-PCR were obtained using throat swab specimens while 34 positive and 38 negative tests for the N-specific tests were available. Similarly, the great majority of negative results (56 from 57 for ORF1ab RT-PCR and 34 from 38 for N-specific RT-PCR) corresponded to RPP30 Ct values above the established cut off values (Fig. 4J). These data demonstrated that the majority of false negative results could be correctly identified by using the established cutoff Ct values for RPP30.
-
In a recent report, recovered COVID-19 patients were repeatedly found positive in SARS-CoV-2 RNA testing during post-discharge monitoring. Based on this observation, the possibility of latent or chronic SARS-CoV-2 infection has been suggested. Contradicting test results from the same patients are not rare in the diagnosis of and clinical care for COVID-19 patients. In fact, the same were observed in some patients enrolled in this study.
In order to evaluate the roles of false negative played in contradicting test results for same patients, we analyzed data for a patient from whom sputum (n = 13) and throat swab (n = 10) specimens were collected at different time points during the course of infection. From sputum specimens, 5 positive and 8 negative RT-PCR results were obtained for the ORF1ab region amplification, while 10 positive and 3 negative results for the N region amplification (Fig. 5A). Three positive and 7 negative RT-PCR results were found for the ORF1ab with throat swab specimens while 4 positive and 6 negative results were obtained for the N region (Fig. 5B). Not surprisingly, negative RT-PCR results for SARS-CoV-2 RNAs corresponded to high Ct values for RPP30 RNAs which were above the established cut off values.

Figure 5. The longitudinal analysis of RT-PCR results of SARS-CoV-2 and RPP30 RNAs in sputum and throat swab specimens from patients with conflicting RT-PCR results. 13 sputum (A) and 10 throat swab (B) specimens were collected from a single patient at multiple time points after disease onset and subjected to SARS-CoV-2 specific RT-PCR assays targeting the ORF1ab and N region and RPP30 RT-PCR. Green lines: cutoffs = 38 for Ct values, positive results < 38. The Ct values of RPP30 RT-PCR set as cutoffs are indicated by black lines (21.53 in A, 25.53 in B) and yellow lines (24.47 in A and 27.48 in B).
When the same analyses were expanded to a cohort of 25 patients who received contradicting test results at different time points, the same correspondence between high Ct values for RPP30 and negative test results was found (Tables 1, 2). Apparently, the detectability of SARS-CoV-2 RNA in these patient specimens was strongly correlated with the Ct values of RPP30 RT-PCR reaction. Thus, conflicting results were mainly caused by false negativity resulted from poor quality of patient specimens. Besides, the conclusion of reactivation of or reinfection with SARS-CoV-2 can be drawn only after false negative results have been excluded.
Patient Test (n) Ct value of RPP30 ≤ 21.53 21.53–24.47 > 24.47 SARS-CoV-2 RNA (N or ORF1ab region) +/+ ± −/− +/+ ± −/− +/+ ± −/− P1 4 1 1 2 P2 6 1 3 2 P3 11 1 3 4 1 2 P4 5 1 2 2 P5 8 3 1 2 2 P6 7 1 3 1 2 P7 7 4 2 1 P8 10 2 3 5 P9 12 2 5 1 2 2 P10 5 2 1 1 1 P11 3 2 1 P12 7 3 2 1 1 P13 7 4 2 1 P14 13 5 5 3 P15 4 1 1 1 1 Total 109 31 1 21 28 3 3 22 Table 1. Correlation between Ct values of RPP30 and detection results of SARS-CoV-2 RNA in sputums
Patient Test (n) Ct value of RPP30 ≤ 25.53 25.53–27.48 > 27.48 SARS-CoV-2 RNA (N or ORF1ab region) +/+ ± −/− +/+ ± −/− +/+ ± −/− W1 7 2 3 2 W2 14 4 1 9 W3 5 1 1 1 2 W4 6 1 2 1 2 W5 6 1 2 3 W6 9 1 1 1 1 1 4 W7 7 1 1 1 4 W8 5 2 1 1 1 W9 10 2 1 1 6 W10 3 1 1 1 Total 72 10 1 4 16 4 1 2 34 Table 2. Correlation between Ct values of RPP30 and detection results of SARS-CoV-2 RNA in throat swabs
RNA Detection in Sputum and Throat Swab Specimens
Relationship among Amplification of Target Regions ORF1ab, N Region and Cellular Gene RPP30
Discrimination of False Negative Results in RT-PCR Detection of SARS-CoV-2 RNAs
Conflicting Results Caused by False Negativity Resulted from Poor Quality Specimens
-
In the present study, we showed that reliable detection of SARS-CoV-2 RNA by RT-PCR assays is clearly associated with the content of cellular materials in patient samples, assessed by the RNA levels of RPP30. A plausible explanation for this finding is that SARS-CoV-2 RNA level may be high in infected epithelial cells and could be more easily detected by RT-PCR assays. This might imply that the detected viral RNA level does not reflect the titer of free viral particles and are not correlated with viral loads. This hypothesis needs to be verified by further examination of the levels of viral RNA in infected host cells and free viral particles though. Nevertheless, the procedures to collect patient specimens need to be performed by trained staffs, ensuring that the specimens contain enough cellular materials.
In recent reports, recovered COVID-19 patients, defined by negative test results for SARS-CoV-2 RNA in at least two independent tests, were found repeatedly positive for SARS-CoV-2 RNA testing during post-discharge monitoring (Chen et al. 2020a). This observation has triggered great concern about the possibility of reactivation or latent/chronic SARS-CoV-2 infection. In addition, contradicting test results from the same patients are not rare in the diagnosis and clinical care for COVID-19 patients. Our retrospective analysis of data for patient samples showed that the negative test results of SARS-CoV-2 were mainly caused by the poor quality of specimens that did not contain sufficient amount of cellular materials.
Our study demonstrated that a reliable result of SARS-CoV-2 RNA test needs an appropriate quality control based on the assessment of cellular markers such as RPP30 RNA levels. Specific cutoffs based on RPP30 RNA detection could be established to evaluate the reliability of SARS-CoV-2 RNA test and identify false negative results. These Ct cutoff values of RPP30 RT-PCR can serve as criteria for identification of false negative results. In clinical application, RPP30 may be integrated in the PCR reactions as an internal reference. In this study, Ct values 21.53 and 25.53 are set as criteria for quality control for ORF1ab region amplification with sputum and throat swab specimens respectively (Fig. 6). Similarly, specific Ct values for quality control purpose should be established on the basis of data collected from tests with specific standardized method and procedure.

Figure 6. Flow chart of SARS-CoV-2 RNA test and evaluation. Sputum and throat swab samples will be evaluated for their suitability for SARS-CoV-2 RNA test based on Ct values of RPP30 RT-PCR. The samples with Ct values of RPP30 RT-PCR below the specific cutoffs will generate clear positive and negative results, while those with a single positive result need to be retested. For the samples with Ct values of RPP30 RT-PCR above the specific cutoffs, either a single positive or double negative result need to be retested.
Our results confirmed that the positive rate of SARS-CoV-2 RNA detection is higher when sputum instead of throat swab specimens (Pan et al. 2020; Yang et al. 2020c). Based on this study, this can be is explained by higher cellular contents in sputum specimens compared with that in throat swabs.
Taken together, a great number of false RT-PCR test results reported are apparently results of inadequate quality of patient specimens and can be excluded by leveraging data for integrated internal reference in the test reactions to assess the content of cellular materials.
-
This work was supported by the Natural Science Foundation of Anhui Province (Grant No. 1608085MH162). We are grateful to Dr. Delia Cosgrove from University of Duisburg-Essen for language editing.
-
ML and ZZ conceptualized and designed the study; YZ, CW, MH, JY and YG performed the experiments; ZL, TH, TL, MX, LZ and GZ collected data and carried out the initial analyses; CW, MH and YG drafted the initial manuscript, YZ, JY, GZ, ML and ZZ reviewed and revised the manuscript. All authors read and approved the final manuscript.
-
The authors declare that they have no conflict of interest.
-
The study was approved by the Ethics Committee of the second hospital of Fuyang (Number 2020020004). Written informed consents were obtained from all patients.
















 DownLoad:
DownLoad: